About ABCMdb
SummaryFuture directions
Regions
Copyrights
Summary
ABC (ATP-Binding Cassette) transporters with altered function are responsible for numerous human diseases. To overcome the pathological phenomena and plan strategies to modulate their function, the mechanism of the action of ABC transporters is studied extensively. The major way to learn about the mechanism of ABC transporters is designing and studying mutant constructs.
Selecting the position and amino acid for mutagenetic experiments raises trivial questions, such as whether or not a particular position has been mutated by others, what type of amino acid has been used to replace the original amino acid, what the environment of that position is in the 3D structure.
To answer these questions, we set up ABCMdb (this database and web application) that contains not only mutations described as disease causing alterations, but also mutations generated for functional studies.
1. Mutations are identified automatically and stored in the database
The MutationFinder software was used to identify mutations in selected full-text papers for several ABC proteins. To decrease the number of false positive hits we developed additional rules and algorithms to complement MutationFinder function, these methods have been wrapped in the MutMiner recognition pipeline.
2. Alignments for comparative analysis of mutations
We generated alignments to identify mutations at similar positions as the mutation of interest in a given ABC protein. For example, you can look up mutations in ABC proteins in the database at analogous positions as Q141K in ABCG2 (Browse – select ABCG2 – list all mutations – click on homologous mutations in the row of Q141K). Since the references to publications describing mutations are also stored in the database, it is effortless to collect comprehensive information on mutations (click on the amino acid in the alignment; or Browse – select ABCG2 – list all mutations – details in the row of Q141K).
Similar to homology model building, this type of comparative analysis is highly sensitive to the alignment used. To allow using custom alignments we provide the possibility to upload your own alignments (Login – MyProfile – alignments).
3. Mapping the mutations in 3D
Visualization of the position of amino acids of interest in 3D can advance experimental design. The database provides the possibility of visualization using available X-ray structures and homology models. By clicking on amino acids in the selected alignment you can toggle the colored display of the given amino acid in the selected structural model (Browse – select a protein – map mutations).
Taking the accuracy of homology models into account, these models advance mutagenic experiments to decipher relationships of structure and mechanism in ABC transporters. Although there are default homology models and X-ray structures built into the application, you also can upload your own structures (Login – MyProfile – PDB files).
Future directions
- Upload and analyze literature data for all human ABC proteins;
- Include non-human ABC proteins in the database;
- Automatic detection of type of the mutations (e.g. disease associated, affecting function or stability);
- Increase user-friendiness
Regions
To help placing a given mutation in logical, functional, and structural contexts, most important regions in ABC transporters are indicated. The trivial sequence parts (non-transmembrane, such as nucleotide binding domains and Walker A regions) were determined by ProSite tools.
The transmembrane helix boundaries were predicted using experimental papers, the HMMTOP server, X-ray structures and the TMDET server and sequence alignments. Please, keep in mind that the accuracy of sequence-based prediction of transmembrane helices in ABC proteins is often erroneous, and the correct determination of transmembrane regions depends heavily on available structures, biochemical data and the quality of sequence alignments.
The naming system for the individual regions is illustrated on a hypothetical super ABC chymera in the figure below, which contains all regions referenced in the database.
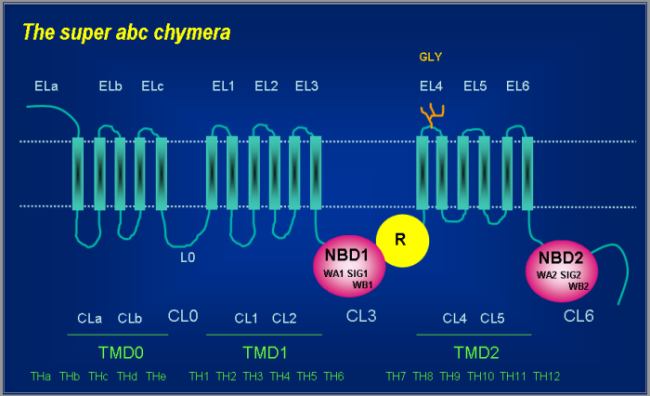
The abbreviations used in the figure are the following:
- TMD: transmembrane domain
- NBD: nucleotide binding domain
- EL: extracellular loop
- CL: cytoplasmic loop
- TH: transmembrane helix
- WA: Walker A motif
- WB: Walker B motif
- SIG: ABC signature sequence (C-motif)
Copyrights
For getpdf and MutationFinder please find copyright notes at the MutMiner site.
We have considered two main points using sentences cited from the literature.
- The policy of many journals: welcome to copy, distribute, transmit and adapt the work (at no cost and without permission) for noncommercial use as long as they attribute the work to the original source using a citation. However, we are not able to check the policy of every journal. If you think we violate the copyright of your publisher, please, contact us.
- Sentences are extracted from their textual and logical environment in a similar way as a 10 sec long melody from a 10 min music. We also think that these sentences will stimulate users to read (purchase) the full text papers.
The database and webapplication were designed and programmed by G. Gyimesi, D. Borsodi, and T. Hegedűs.
Manual verification and correction of data to enhance MutMiner were done by H. Tordai and H. Sarankó.
Thanks for useful suggestions and comments to Paula Duek Roggli (Bairoch Group, Swiss Institute of Bioinformatics).
